OPERATE/SPECTRUM result_spectrum = spectrum_1 operation spectrum_2Add, subtract, divide, or time-average (operation:
SIMULATE/SPECTRUM par [bin] [out] [efa] [drm] [cts] [time] [seed]Simulate a spectrum. A model must be defined in parameter file par by using the command CREATE/PARFIL fit. Parameter bin needs the name of a table (default: phcgrid) which contains a binned grid of pulse height channels (column labels L_CH and H_CH for lowest and highest channel). It must have a general EXSAS header with file type SPE. The simulated spectrum will be output to out - (default: <bin>_o). Instead of the usual correction table the effective area table (default: exsas_cal:effarea_pspcb) has to be used. The detector response matrix (default: exsas_cal:drm<detector_id>) can optional be given to parameter drm. The next three parameters are the number of counts (default value = 1000), the observation time in seconds (default value = 1000), and a negative seed number to start the random number generator (default value = -17777).
CREATE/PARFIL FIT parameter_file ? [parameter] [value] [expert_flag]Define a parameter_file for MODEL/SPECTRUM. A parameter and its value may be inserted in the parameter file while creating it. If no expert_flag is given key words are explained in full detail.
BIN/DETECTOR observed_spectrum [input_drm] [output_drm]Bin the original input detector response matrix drm according to the binning of the observed spectrum and store the result in the output file.
Note:The default for input depends on your observed spectrum and is always the response matrix which is best suited for your observation. It may change in future EXSAS releases as calibration improves.
MODEL/SPECTRUM parameter_file [observed_spectrum] [model_result]
[correction_table] [detector_response_matrix]
Model a spectrum according to specifications given
in the parameter file. As options the observed
spectrum, an output file, a correction table,
and a detector response matrix can be given.
FIT/SPECTRUM parfile/acronym intab [outtab] [cortab] [drmat] [plot]Fit a model (specified in parfile or as 4-letter acronym) to an observed spectrum given as intab and output the result into table outtab by using the correction vector cortab and the detector response matrix drmat. The option plot allows to plot the result without calling explicitly the command PLOT/FIT.
Note:This command combines BIN/DETECTOR_RESPONSE, MODEL/SPECTRUM, and PLOT/FIT. The defaults for cortab and drmat depend on your observed spectrum intab and may change in future EXSAS releases. File names defined in a parameter file are always overwritten by default names or names defined on the command line. Of course, the four letter acronym is a preserved name for a parameter file and cannot be used in this command as a user defined parameter file name.
INTEGRATE/FLUX input_file E_min E_max [frame] [H0] [q0]Integrate a flux density given in an input_file, which must be an output file of MODEL/SPECTRUM, between the energy boundaries of E_min and E_max. For a red-shifted source also a luminosity is calculated. If no options are used, the defaults are: frame = SOURCE, H0 = 50km/s/Mpc, q0 = 1/2.
CONVERT/RATE_TO_FLUX par_file rate/tbl detector [E_low] [E_high]The accepted command line options are: par_file is a parameter file in the usual EXSAS format which contains the spectral model and its parameters. Important to note is that the program assumes always that in par_file PARAMETER 1 is the column density and PARAMETER 2 is the photon flux amplitude in photons/s/cm2. An example of a valid parameter file is:
!
! TASK: FIT
!
! --- power law ---
MODEL gamm(1)*powl(2,3,4) ! gal_abs.*power_law
PARAMETER 1 0.08 ! x 10^21 cm^-2
PARAMETER 2 1.0 ! photon flux ampl. at E0
PARAMETER 3 -2.6 ! photon index
PARAMETER 4 1.0 ! ref_energy E0 keV
NOTE: The line "! TASK: fit" is mandatory
The required parameter file has to define only the spectral
model and the "Task" which is "FIT". No additional directives
are needed.
Note:This command does not support models which are normalized to the integrated energy flux or normalized to luminosity flux, i.e. don't use models like BBEF, PLEF, BPEF, EXEF, EPEF, TBEF, GSEF, BBLM, PLLM, BPLM, EXLM, EPLM, TBLM to convert the count rate to the integrated energy flux. Also care has to be taken using user defined models (!).The command works fine with the additive models: BBDY, DIBB, POWL, BRPL, PWLB, EXPO, PLEX, THBR, COMP, RSMF, GAUS and ASYG.
Another point to note is that always the instrument's on-axis effective area is used during the energy flux computation. This is not a restriction if you have deduced your count rate with one of the EXSAS spatial analysis programs. However, if your source is not on-axis and you have used some non-EXSAS procedure for the count rate estimate make shure that your count rate is vignetting corrected (!).
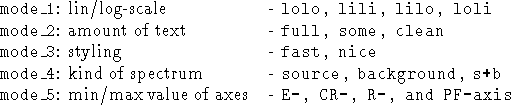
PLOT/FIT_SPECTRUM input_file [mode_1] [mode_2] [mode_3] [mode_4]
[mode_5]
Fitted data and observed data from
input_file are plotted according
to four plot modes (first options = defaults):

Note:In order to avoid an unexpected bad overplotting of a spectrum which occurs if one of the plotted columns has a zero or even negative value a selection of the table according to the specific column is suggested.
PLOT/ERROR_ELLIPSE input_file fit_par1 fit_par2 [sc_mode] [cl_mode]Error ellipses of covariance matrix from input file are plotted for two given fit parameters according to scale mode (Y/N, default=Y) and mode of confidence levels (Decimals/Sigmas, default=S).
PLOT/CHI2_CONTOUR frame [ncont] [chimin] [chicon] [smpar] [xlab] [ylab]One or two dimensional
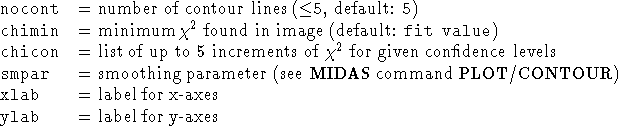
Note:For a better quality of this plot one has to apply the full capacity of the Midas plotting commands.
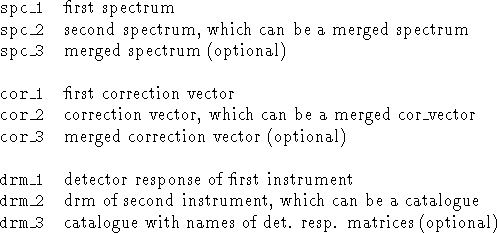
MERGE/SPECTRA spc_1,spc_2[,spc_3] cor_1,cor_2[,cor_3] drm_1,drm_2[,drm_3]This command has three lists of names where each list consists of two or three file names separated by commas. The first list contains the names of the spectra, the second list the names of their corresponding correction vectors and the third the names of the corresponding detector response matrices. No default names are defined.
TUTORIAL/SPECTRAL_ANALYSIS [observed_spectrum_file] [expert_flag]The file with the observed spectrum and its corresponding correction vector must be present in the working directory, if not the default name (spectrum) is used. In this optional case one has to update both parameter files, which are opened during the session, by command CREATE/PARFIL fit. Each command of this tutorial will be explained in detail, if no expert flag (any string) is given.