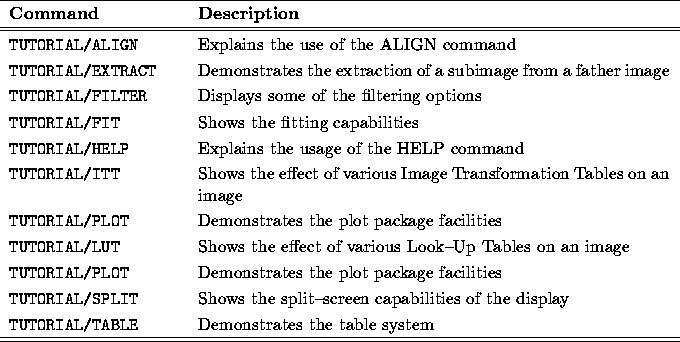
Table: List of Tutorials
This section gives two examples of simple MIDAS sessions. The first one reads some frames from a magnetic tape, displays them on a monitor and performs some simple operations. The second one creates MIDAS tables and performs some simple operations. In the following examples, user input is written in bold face type while comments, (after an exclamation mark) are written in normal roman font.
$ inmidas
Midas 001> HELP INTAPE !get help for tape input
Midas 002> INTAPE * x TAPE1 FNN !lists headers of all files
!from tape mounted on TAPE1
Midas 003> INTAPE 2,16-17,31-33,52 CCD TAPE1 !read files from TAPE1
Image ccd0002 : FF D V 20S , naxis: 2, pixels: 337, 520 Image ccd0016 : DK BIAS 1S , naxis: 2, pixels: 337, 520 Image ccd0017 : DK BIAS 1S , naxis: 2, pixels: 337, 520 Image ccd0031 : DK 60S , naxis: 2, pixels: 337, 520 Image ccd0032 : DK 60S , naxis: 2, pixels: 337, 520 Image ccd0033 : DK 60S , naxis: 2, pixels: 337, 520 Image ccd0052 : A0532-527 V 300 , naxis: 2, pixels: 337, 520Midas 004> INTAPE 78 ccd image.fits !read fits file from disk
Image ccd0078 : A1029-459 V 40S , naxis: 2, pixels: 337, 520Midas 005> CREATE/ICAT !create a catalogue of images
Image catalog icatalog.cat with 8 entries created...
Midas 006> SET/ICAT !enable catalogue
Midas 007> READ/ICAT !list the catalogue out
Image Catalog: icatalog.catNo Name Ident Naxis Npix(1,2) #0001 ccd0002.bdf FF D V 20S 2 337 520 #0002 ccd0016.bdf DK BIAS 1S 2 337 520 #0003 ccd0017.bdf DK BIAS 1S 2 337 520 #0004 ccd0031.bdf DK 60S 2 337 520 #0005 ccd0032.bdf DK 60S 2 337 520 #0006 ccd0033.bdf DK 60S 2 337 520 #0007 ccd0052.bdf A0532-527 V 300 2 337 520 #0008 ccd0078.bdf A1029-459 V 40S 2 337 520Midas 008> STAT/IMA ccd0016 !compute statistics for this frame
frame: ccd0016 (data = R4) complete area of frame minimum, maximum: 184.0000 16383.00 at ( 215, 2),( 337, 520) mean, standard_deviation: 251.2245 880.0140 3rd + 4th moment: 0.1305419E+11 0.2137182E+15 total intensity: 0.4402457E+08 median, 1. mode: 16287.41 16287.71 total no. of bins, binsize: 256 63.52549 # of pixels used = 175240 or 100.00 % of all possible pixels (= 175240) from 1 1 to 337 520 (in pixels)Midas 009> STAT/IMA ccd0017frame: ccd0017 (data = R4) complete area of frame minimum, maximum: 187.0000 16383.00 at ( 113, 1),( 337, 520) mean, standard_deviation: 251.2514 880.0129 3rd + 4th moment: 0.1305419E+11 0.2137182E+15 total intensity: 0.4402929E+08 median, 1. mode: 16287.30 16287.73 total no. of bins, binsize: 256 63.51373 # of pixels used = 175240 or 100.00 % of all possible pixels (= 175240) from 1 1 to 337 520 (in pixels)Midas 010> CREATE/DISPLAY !create a display window
Midas 011> LOAD/IMA ccd0017 !display image
Midas 012> GET/CURS !read some pixels values from the image
cursor #0 frame pixels world coords intensity frame: ccd0017 334.0 166.0 334.000 166.000 228.000Midas 013> EXTRACT/IMA ff = ccd0002[<,<:@330,>] !remove
!irrelevant columns
Midas 014> EXTRACT/IMA biai1 = ccd0016[<,<:@330,>]
Midas 015> EXTRACT/IMA biai2 = ccd0017[<,<:@330,>]
Midas 016> EXTRACT/IMA dk1 = ccd0031[<,<:@330,>]
Midas 017> EXTRACT/IMA dk2 = ccd0032[<,<:@330,>]
Midas 018> EXTRACT/IMA dk3 = ccd0033[<,<:@330,>]
Midas 019> EXTRACT/IMA ima1 = ccd0052[<,<:@330,>]
Midas 020> EXTRACT/IMA ima2 = ccd0078[<,<:@330,>]
Midas 021> STAT/IMA biai1frame: biai1 (data = R4) complete area of frame minimum, maximum: 184.0000 497.0000 at ( 215, 2),( 147, 408) mean, standard_deviation: 203.2299 3.752572 3rd + 4th moment: 8402620. 0.1709546E+10 total intensity: 0.3487425E+08 median, 1. mode: 202.0980 204.2529 total no. of bins, binsize: 256 1.227451 # of pixels used = 171600 or 100.00 % of all possible pixels (= 171600) from 1 1 to 330 520 (in pixels)Midas 022> STAT/IMA biai2frame: biai2 (data = R4) complete area of frame minimum, maximum: 187.0000 613.0000 at ( 113, 1),( 286, 473) mean, standard\_deviation: 203.2554 3.836732 3rd + 4th moment: 8406459. 0.1710918E+10 total intensity: 0.3487864E+08 median, 1. mode: 201.6387 202.8706 total no. of bins, binsize: 256 1.670588 # of pixels used = 171600 or 100.00 % of all possible pixels (= 171600) from 1 1 to 330 520 (in pixels)Midas 023> READ/DESCR biai1 STATISTIC !read descriptor statistic
frame: BIAI1 (data = R4) STATISTIC : 184.0000 497.0000 203.2299 3.752572 8402620. 0.1709546E+10 202.0980 204.2529 256.0000 1.227451 0.3487425E+08Midas 024> COMPUTE/IMA ffb = ff-203. !biais correction
Midas 025> COMPUTE/IMA dk1b = dk1-203.
Midas 026> COMPUTE/IMA dk2b = dk2-203.
Midas 027> COMPUTE/IMA dk3b = dk3-203.
Midas 028> COMPUTE/IMA ima1b = ima1-203.
Midas 029> COMPUTE/IMA ima2b = ima2-203.
Midas 030> AVERAGE/IMA dk = dk1b,dk2b,dk3b !compute an
!average of DARK frames
dk1b processed ...
dk2b processed ...
dk3b processed ...
Midas 031> COMPUTE/IMA ffd = ff-dk !dark subtraction
Midas 032> COMPUTE/IMA ima1bd = ima1b-dk
Midas 033> COMPUTE/IMA ima2bd = ima2b-dk
Midas 034> COMPUTE/IMA ima1bdf = ima1bd/ffd !flat-field the frame
Midas 035> COMPUTE/IMA ima2bdf = ima2bd/ffd
Midas 036> LOAD/IMA ima1bdf
Midas 037> READ/DESCR ima1bdf LHCUTSframe: ima1bdf (data = R4) LHCUTS : 0.0000000E+00 0.0000000E+00 -0.7500000E-01 1.992324 0.0000000E+00 0.0000000E+00Midas 038> CUTS ima1bdf 0.,1.992 !modify display cuts
Midas 039> LOAD ima1bdf
Midas 040> READ/ICAT icatalog
Image Catalog: icatalog.catNo Name Ident Naxis Npix(1,2) #0001 ccd0002.bdf FF D V 20S 2 337 520 #0002 ccd0016.bdf DK BIAS 1S 2 337 520 #0003 ccd0017.bdf DK BIAS 1S 2 337 520 #0004 ccd0031.bdf DK 60S 2 337 520 #0005 ccd0032.bdf DK 60S 2 337 520 #0006 ccd0033.bdf DK 60S 2 337 520 #0007 ccd0052.bdf A0532-527 V 300 2 337 520 #0008 ccd0078.bdf A1029-459 V 40S 2 337 520 #0009 ff FF D V 20S 2 330 520 #0010 biai1 DK BIAS 1S 2 330 520 #0011 biai2 DK BIAS 1S 2 330 520 #0012 dk1 DK 60S 2 330 520 #0013 dk2 DK 60S 2 330 520 #0014 dk3 DK 60S 2 330 520 #0015 ima1 A0532-527 V 300 2 330 520 #0016 ima2 A1029-459 V 40S 2 330 520 #0017 ffb FF D V 20S 2 330 520 #0018 dk1b DK 60S 2 330 520 #0019 dk2b DK 60S 2 330 520 #0020 dk3b DK 60S 2 330 520 #0021 ima1b A0532-527 V 300 2 330 520 #0022 ima2b A1029-459 V 40S 2 330 520 #0023 dk average frame 2 330 520 #0024 ffd FF D V 20S 2 330 520 #0025 ima1bd A0532-527 V 300 2 330 520 #0026 ima2bd A1029-459 V 40S 2 330 520 #0027 ima1bdf A0532-527 V 300 2 330 520 #0028 ima2bdf A1029-459 V 40S 2 330 520Midas 041> OUTTAPE icatalog,9-28 TAPE1 !save data on tape
File ff.bdf written to tape with 241 blocks File biai1.bdf written to tape with 243 blocks File biai2.bdf written to tape with 243 blocks File dk1.bdf written to tape with 241 blocks File dk2.bdf written to tape with 241 blocks File dk3.bdf written to tape with 241 blocks File ima1.bdf written to tape with 241 blocks File ima2.bdf written to tape with 241 blocks File ffb.bdf written to tape with 241 blocks File dk1b.bdf written to tape with 241 blocks File dk2b.bdf written to tape with 241 blocks File dk3b.bdf written to tape with 241 blocks File ima1b.bdf written to tape with 241 blocks File ima2b.bdf written to tape with 241 blocks File dk.bdf written to tape with 241 blocks File ffd.bdf written to tape with 241 blocks File ima1bd.bdf written to tape with 241 blocks File ima2bd.bdf written to tape with 241 blocks File ima1bdf.bdf written to tape with 241 blocks File ima2bdf.bdf written to tape with 241 blocksMidas 042> OUTTAPE icatalog,7-8 ima !save data on disk in FITS format
File ima1.bdf written to disk> ima0001.mt File ima2.bdf written to disk> ima0002.mtMidas 043> PRINT/LOG !print logfile on the default printer
Midas 042> BYE
$ inmidas
Midas 001> CREATE/TABLE flux1 2 30 flux1 ! create table from ASCII file
Midas 002> SHOW/TABLE flux1 ! list table parameters
Table : flux1 [Transposed format] No.Columns : 2 No.Rows : 30 All.Columns: 3 All.Rows : 32 Sorted by Sequence Reference : Sequence Col.# 1:LAB001 Unit:Unitless Format:E15.6 R*4 Col.# 2:LAB002 Unit:Unitless Format:E15.6 R*4 Selection : ALLMidas 003> READ/TABLE flux1 :LAB001 :LAB002 @1 @9
!read data from table
Table : flux1 Sequence LAB001 LAB002 -------- --------------- --------------- 1 4.71000e+03 1.12850e+04 2 4.77000e+03 1.08300e+04 3 5.05000e+03 8.77000e+03 4 5.09000e+03 8.55000e+03 5 5.12000e+03 8.34000e+03 6 5.19000e+03 7.93000e+03 7 5.23500e+03 7.73500e+03 8 5.26500e+03 7.55000e+03 9 5.39000e+03 7.00500e+03 -------- --------------- ---------------Midas 004> NAME/COLUMN flux1 :LAB001 "(sec)" F6.0 !change
! format and define unit
Midas 005> NAME/COLUMN flux1 :LAB002 "(erg/sec)" F6.0
Midas 006> READ/DESCR flux1.tbl HISTORY
HISTORY : CREA/TABL flux1 2 30 flux1 NULL TRAN NAME/COLU flux1 :LAB001 "(sec)" F6.0 NAME/COLU flux1 :LAB002 "(erg/sec)" F6.0Midas 007> STAT/TABLE flux1 :LAB001 !compute statistics of
!one of the column
Table : flux1 Column # 1 Label :LAB001 Type :R*4 Total no. of entries : 30, selected no. of entries : 30 Miminum value : 0.47100E+04, Maximum value: 0.96800E+04 Mean value : 0.66437E+04, Standard dev.: 0.15539E+04Midas 008> $more flux2.fmt! ! format file flux2.fmt ! DEFINE/FIELD 1 6 R F6.0 :LAB001 DEFINE/FIELD 9 13 R F6.0 :LAB002 ENDMidas 009> CREATE/TABLE flux2 2 8 flux2 flux2
Midas 010> READ/TABLE flux2Table : flux2 Sequence LAB001 LAB002 -------- ------ ------ 1 9705 2750 2 9930 2700 3 10130 2655 4 10150 2650 5 10520 2580 6 10740 2540 7 11040 2485 8 11570 2410 -------- ------ ------Midas 011> MERGE/TABLE flux1 flux2 flux !merge of two tables
Midas 012> SHOW/TABLE fluxTable : flux [Transposed format] No.Columns : 2 No.Rows : 38 All.Columns: 3 All.Rows : 40 Sorted by # Sequence Reference : Sequence Col.# 1:LAB001 Unit:(sec) Format:F6.0 R*4 Col.# 2:LAB002 Unit:(erg/sec) Format:F6.0 R*4 Selection : ALLMidas 013> SORT/TABLE flux :LAB002 !sort table according to
!increasing values of a column
Midas 014> REGRESSION/POLYNOMIAL flux :LAB001 :LAB002 5 !polynomial fit
flux POLYNOMIALS Input Table : UNION Type : MUL L-S N.Cases : 38 ; N.Ind.Vars : 1 Dependent variable : column # 1 Independent variable: column # 2 degree : 5 degree 0 4.4818E+04 1 -2.6778E+01 2 7.4671E-03 3 -1.0405E-06 4 7.1309E-11 5 -1.9126E-15 R.M.S error : 72.04152Midas 015> SAVE/REGRESSION flux REGRE !save the result of
!regression in a descriptor
Midas 016> CREATE/COLUMN flux :FIT !create a new column
!in the table
Warning: Column overflow mechanism activated
Midas 017> COMPUTE/REGRESSION flux :FIT = REGRE !compute
!the results of the regression
Midas 018> READ/TABLE flux :FIT @1..4,@6,@9Table : flux Sequence FIT -------- ------------- 1 1.1337231e+04 2 1.0955642e+04 3 1.0690593e+04 4 1.0505399e+04 6 1.0174703e+04 9 9.7462227e+03 -------- -------------Midas 019> CREATE/GRAPH
!create graphic window
Midas 020> PROJECT/TABLE flux newflux :LAB002 ! project one
!column of a table in a new one
Midas 021> INTERPOLATE/TT newflux :LAB002,:SPLINE flux :LAB002,:LAB001 0.001
!spline interpolation
Midas 022> CREATE/GRAPH
!create graphic window
Midas 023> PLOT/TABLE newflux :LAB002 :LAB001 !plot table columns
Midas 024> SET/PLOT LTYPE=1 STYPE=0 !plot table columns
Midas 025> OVERPLOT/TABLE flux :LAB002 :SPLINE
Midas 026> SELECT/TAB newflux sequence.gt.5 !select part of the table
Midas 027> COPY/TAB newflux result !copy selected table
Midas 028> OUTTAPE result.tbl result.fits !save file in FITS
!format on disk
Midas 029> BYE